Galaxy@UMBI-HPC: The Answer to Omics Big Data

Ang Mia Yang
Associate Professor Dr. Neoh Hui-min
According to Techopedia, big data refers to a collection of data sets, so large and complex that it becomes difficult to process using on-hand database management tools or traditional data processing applications. The challenges of big data include capture, curation, storage, search, sharing, transfer, analysis and visualization processes.
As the cost of sequencing decreases, it will become easier to generate huge amounts of data. Rapid technology development means applications and software have to keep up with ever-growing volume and numerous data characteristics. Both features encourage a centralized approach to data storage and analysis.
We would like to introduce Galaxy@UMBI-HPC, a bioinformatics workflow management system for modern omics research. Aimed to increase access to complex computational analyses for all scientists, including those with limited or no programming knowledge, Galaxy@UMBI-HPC is established with a web-based graphical user interface (GUI), making it simple to do everything needed for data analyses.

Galaxy@UMBI-HPC runs on our Linux-based server, where end users will be able to access it from any computer operating system equipped with up-to-date web browsers. Paired with our high-performance computer cluster, the UMBI-HPC, all individual tools offered are installed beforehand, meaning the end user does not have to download or install the tools themselves. This avoids various practical problems associated with deployment and update of tools to individual desktops – among them are complications in installation, differences between versions, reliance on unstated dependencies, or in some cases, where these tools may not even run properly on the operating system.
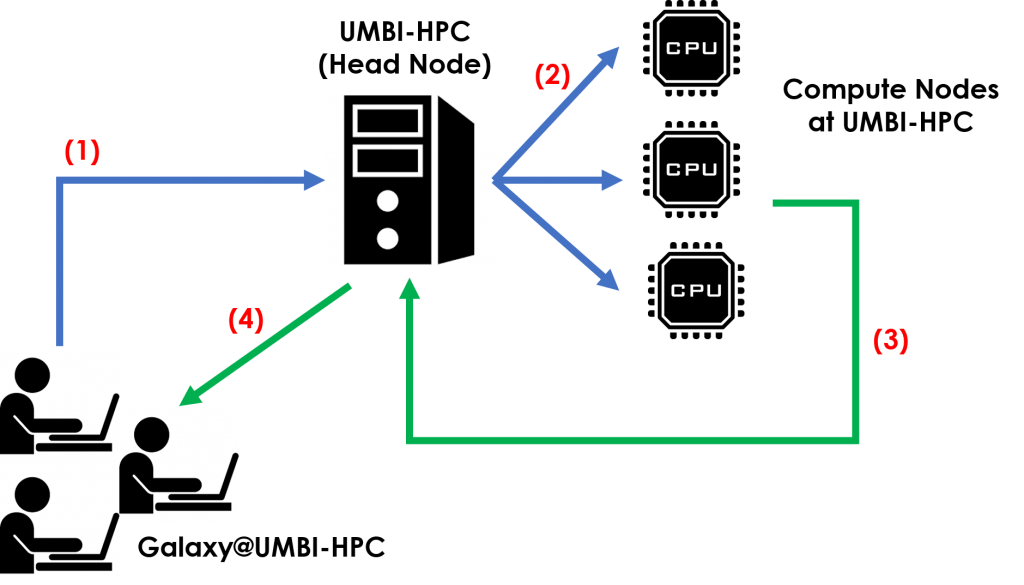
With the GUI, end users can upload their own data or retrieve them from public databases, choose from a plethora of analysis tools, set customized inputs and parameters and run selected tools. The GUI also includes a workflow editor, where users can create automated, multi-step analyses by just drag and drop within fingertips. All results from analyses are completely reproducible, where the inputs, parameters and workflow are permanently recorded, and every analysis can be precisely repeated.
For resources and user support, please refer to our lab manager (umbi.cto@gmail.com).

