Caenorhabditis elegans: a Model Organism for Research
Caenorhabditis elegans: a Model Organism for Research
Mohd Azrul Hisham Ismail
Introduction
Caenorhabditis elegans nematodes are free living organisms. They are very small in size and although it is possible for us to discern the worms using our naked eyes, we need the aid of microscopes to observe the details of C. elegans. [1] Newly hatched larvae are about 0.25 mm long, while the adults are about 1 mm in length.
The worms exist in either male or hermaphrodites forms. Hermaphrodites are able to self-reproduce, and they can do this easily! Males and hermaphrodites are distinguishable through their anatomy. Figure 1 shows the anatomy of hermaphrodites and male C. elegans.
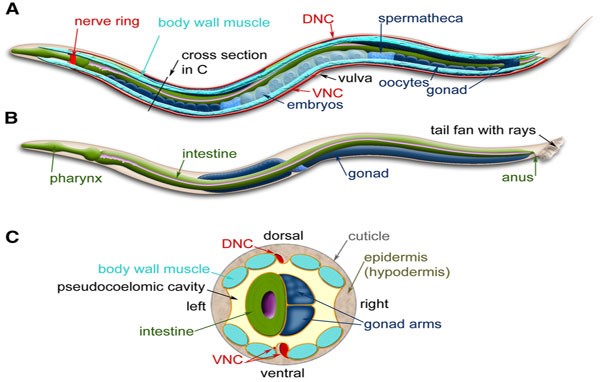
Figure 1: C. elegans anatomy. Major anatomical features of a hermaphrodite (A) and male (B) viewed laterally; (C) Cross-section through the anterior region of the C. elegans hermaphrodite [1]
Hermaphrodites have two ovaries, an oviduct, a spermatheca and a single uterus whereas male nematodes have a single-lobed gonad, a vas deferens and a tail specialized for mating [2]. The easiest way to distinguish between male and hermaphrodites nematodes under a dissecting microscope is to observe its tails. Hermaphrodites have sharp end tails, whereas male nematodes will have tail fans with rays.
Life cycle
C. elegans life cycle consists of different stages. It starts with the embryonic egg stage, four larval stages L1, L2, L3 and L4, and the last stage which is the adult stage. Figure 2 shows the life cycle of C. elegans. Each complete life cycle of C. elegans require a short period of time as short as 3 days [3].
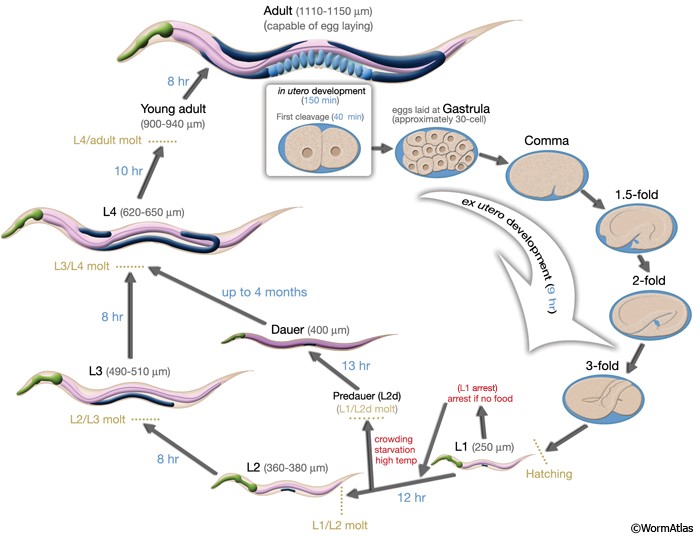
Figure 2: Life cycle of C. elegans [4]
Although, C. elegans is small in size, this organism possesses special characteristics that help them survive in unideal conditions such as lack of food supplies, overpopulation, or extreme temperatures. L2 larvae are able to develop into the “dauer” stage which is an alternative stage in life that permits arrested development in L3 [5]. This upsurge its survival despite lack in essential needs.
Special features of C. elegans for Research Purposes
C. elegans have several special features which make it a model organism for biology research:
1. Genetic similarities
Like other worms, C. elegans do not have complex system or body parts, but, interestingly, they share many genes and molecular pathways with humans. In addition, the complete genome sequence for C. elegans has been published in 1998 [6].
2. Cost effective
C. elegans can be grown at a lower cost compared to other animals or organisms. They only require bacteria specifically E. coli strains OP50 as nutrients for them to survive. They do not require expensive laboratories to grow and can be cultivated in usual laboratories that are equipped with incubators.
3. Easily to maintain
C. elegans are easy to maintain due to their size. No large laboratory space is required to grow them! In addition, nematodes can be frozen and stocked. Healthy cultures of C. elegans can be revived from the frozen stock when required. C. elegans also can reproduce easily. [2] Number of eggs laid by C. elegans can exceed 1,000 when they were inseminated by a male, while up to 300 eggs laid when they were self-inseminated.
C. elegans in Biological Research
Usage of C. elegans for research purposes in biology has increased rapidly. This is due to its features such as rapid generation time with an invariant cell line. There are also well developed genetic and molecular tools for its manipulation. Its simple growth conditions and being the first multicellular animal with complete genome sequence makes it a model organism for biology [7].
A lot of remarkable researches using C. elegans have been published. Notable scientists have been awarded with the Nobel Prize for their outstanding works with the worms. Sydney Brenner, H. Robert Horvitz and John Sulston were awarded with Nobel Prize in Physiology or Medicine in 2002 for their work on genetics of organ development and programmed cell death, meanwhile Andrew Fire and Craig Mello were awarded in 2006 for their discovery of RNA interference in C. elegans. Martin Chalfie with his work on green fluorescent protein (parts of his research using C. elegans) shared a Nobel Prize in Chemistry in 2008 [3].
C. elegans has also been used in neuronal [8], genomic [9], microbiome [10], metabolic diseases [11] and host-pathogen studies [12]. Interestingly, C. elegans has also been to space [13]! It is indeed a useful model organism to be studied.
C. elegans Research in UMBI
C. elegans has been used as a model organism in UMBI’s space and simulated microgravity research [14, 15]. It is now currently being used as a model to investigate host-pathogen interaction in methicillin-resistant Staphylococcus aureus infections.
References
[1] A. K. Corsi, B. Wightman, and M. Chalfie, “A Transparent window into biology: A primer on Caenorhabditis elegans,” in WormBook, 2015, pp. 1–31.
[2] N. Bernabo, I. Saponaro, M. Mattioli, and B. Barboni, “Signaling strategy in Spermatozoa activation of sea urchin, C. elegans and human: three different players for the same melody,” J. Bioeng. Biomed. Sci., vol. s3, no. 001, pp. 1–6, 2012.
[3] D. Dutta and R. Saha, “Contribution of Caenorhabditis elegans in cell cycle research: research needs and research leads,” NBU J. Anim. Sc, vol. 10, pp. 39–47, 2016.
[4] Z. F. Altun and D. H. Hall, “Introduction,” in WormAtlas, 2009.
[5] C. G. James, O. Morah, V. Panwala, and A. Yarmand, “Survival of Caenorhabditis elegans infected with Escherichia coli DFB1655 is not affected by a Missense Mutation in dop-1 or treatment with Chlorpromazine Hydrochloride,” J. Exp. Microbiol. Immunol., vol. 4, no. August, pp. 1–9, 2018.
[6] T. R. Burglin, E. Lobos, and M. L. Blaxter, “Caenorhabditis elegans as a model for parasitic nematodes,” Int. J. Parasitol., vol. 28, pp. 395–411, 1998.
[7] C. L. Kurz and J. J. Ewbank, “Caenorhabditis elegans for the study of host–pathogen interactions,” Trends Microbiol., vol. 8, no. 3, pp. 142–144, 2000.
[8] M. Lakso et al., “Dopaminergic neuronal loss and motor deficits in Caenorhabditis elegans overexpressing human α-synuclein,” J. Neurochem., vol. 86, no. 1, pp. 165–172, 2003.
[9] S. Lall et al., “A genome-wide map of conserved MicroRNA targets in C. elegans,” Curr. Biol., vol. 16, no. 5, pp. 460–471, 2006.
[10] P. Dirksen et al., “The native microbiome of the nematode Caenorhabditis elegans: Gateway to a new host-microbiome model,” BMC Biol., vol. 14, no. 38, pp. 1–16, 2016.
[11] S. Hashmi et al., “A C. elegans model to study human metabolic regulation,” Nutr. Metab., vol. 10, no. 1, pp. 1–11, 2013.
[12] K. M. Balla and E. R. Troemel, “Caenorhabditis elegans as a model for intracellular pathogen infection,” Cell. Microbiol., vol. 15, no. 8, pp. 1313–1322, 2013.
[13] Y. Zhao et al., “A mutational analysis of Caenorhabditis elegans in space,” Mutat. Res. – Fundam. Mol. Mech. Mutagen., vol. 601, no. 1–2, pp. 19–29, 2006.
[14] S. The et al. “Multi-Generational Culture of C. elegans on a Long-Term Space Flight Revealed Changes in Expression of Genes Involved in Longevity, DNA Repair, and Locomotion,” Asia. Pac. J. Mol. Med. vol 4, no. 2, article 1.
[15] L. Tee et al. “Effects of simulated microgravity on gene expression and biological phenotypes of a single generation Caenorhabditis elegans cultured on 2 different media,” Life. Sci. Space Res. (Amst). vol. 15, pp.11-17.

