Kinobeads Profiling To Study Kinase Inhibitor: A Chemical Proteomic Approach For Investigating Drug-Protein Interactions
Written by: Dr. Lee Pey Yee
Date: 19 July 2022
Drug–protein interactions are key to understanding pharmacological therapeutic effects and pleiotropic activity. A comprehensive investigation of drug-protein interactions entails assaying all proteome proteins for drug binding and assessing their affinity for the drug target. For large-scale investigation of protein interactions, several protein ligand-binding assays exist, such as the yeast two-hybrid assay and affinity chromatography coupled with mass spectrometry. In this article, we describe a chemical proteomics technique for detecting drug–protein interactions by using kinobeads profiling of kinase inhibitors as an example.
Due to their crucial function in cellular signalling and their frequent dysregulation in cancer, kinases are among the most popular therapeutic targets nowadays [1]. There are a growing number of targeted small molecule kinase inhibitors that have been developed throughout the past 20 years, most of them for oncological indication [2]. The ATP region is the most common binding site of the kinase inhibitors, however due to sequence conservation within the kinase family, designing highly selective kinase inhibitors is extremely difficult. Although kinase inhibitors are typically regarded as selective agents, it has been found that they can bind to several targets and block numerous pathways simultaneously [3]. The target selectivity of kinase inhibitors might differ greatly. Lapatinib, for instance, only inhibits EGFR and ERBB2 [4], but dasatinib could target approximately 30 tyrosine and serine/threonine kinases [5]. The mode(s) of action of numerous kinase inhibitors are still poorly understood, despite their wide application in clinical practice. Since the drugs have the potential to affect a variety of cellular signalling processes that may cause toxicity and unintended side effects, it is therefore crucial to characterize the target profiles of the kinase inhibitors. On the other hand, kinase off-target findings may reveal unexpected targets that could offer opportunities for drug repurposing.
Kinase panels based on recombinant proteins are often used as the method for drug target profiling [6]. However, these assays are unable to capture the molecular complexity of the target proteins in the cells. Moreover, recombinant proteins do not always resemble natural full-length proteins, which might be post-translationally altered and integrated into protein complexes, which further hinders the accurate identification of drug targets. Chemical proteomics is a powerful method that can overcome many of the shortcomings of the recombinant protein assays to identify the direct protein-ligand interactions [7]. In a typical experiment, target proteins are preferentially enriched by incubation with a cell lysate using a probe compound that is fixed on a solid substrate. Specific targets may be identified by competitive binding with excess free inhibitors, and the binding strength can be assessed through dose-dependent competition experiments that measure the decrease in target binding to the bead matrix as a function of free inhibitor concentration. Finally, the identification of all specifically bound proteins in a single experiment is made possible by the subsequent isolation of the beads and mass spectrometric analysis.
In 2007, Bantscheff and co-workers developed a unique chemical proteomics technique to study the interactions of small molecules compounds with endogenously produced protein kinases [8]. They created “kinobeads,” an affinity resin containing immobilized non-selective kinase inhibitors as a capture tool. The technique involves treating live cells or cell extracts with an inhibitor substance before incubating them with the kinobeads to perform a competitive binding experiment (Figure 1). This approach has been applied to quantitatively profile the kinase inhibitors imatinib, dasatinib, and bosutinib, which verified known drug targets such as ABL and Src family kinase, as well as discovered the receptor tyrosine kinase DDR1 and the oxidoreductase NQO2 as novel imatinib targets [8]. One major advantage of the kinobeads approach for kinase inhibitor profiling is that less studied or unexpected targets of the entire proteome can be identified in an unbiased manner. Furthermore, this technique enables the evaluation of the selectivity and affinity of small molecule inhibitors against endogenously expressed targets in cell extracts or live cells that more closely resemble the native biological condition. Nevertheless, the kinobeads technique still has several limitations. It may miss specific kinases that are not expressed or are only weakly expressed in the cell lines, and it may not capture kinases that have an intrinsic affinity for the beads. Furthermore, the kinobeads assay is restricted to evaluating the binding of targets that are competitive with the ligands; hence, allosteric inhibitor that bind to the site outside the ATP binding pocket cannot be profiled using this technique [9]. This necessitates the need for additional development and refining of the technique. On the other hand, it may be advantageous to combine kinobeads assay with other approaches in an integrated strategy to give complementary information on the drug targets.
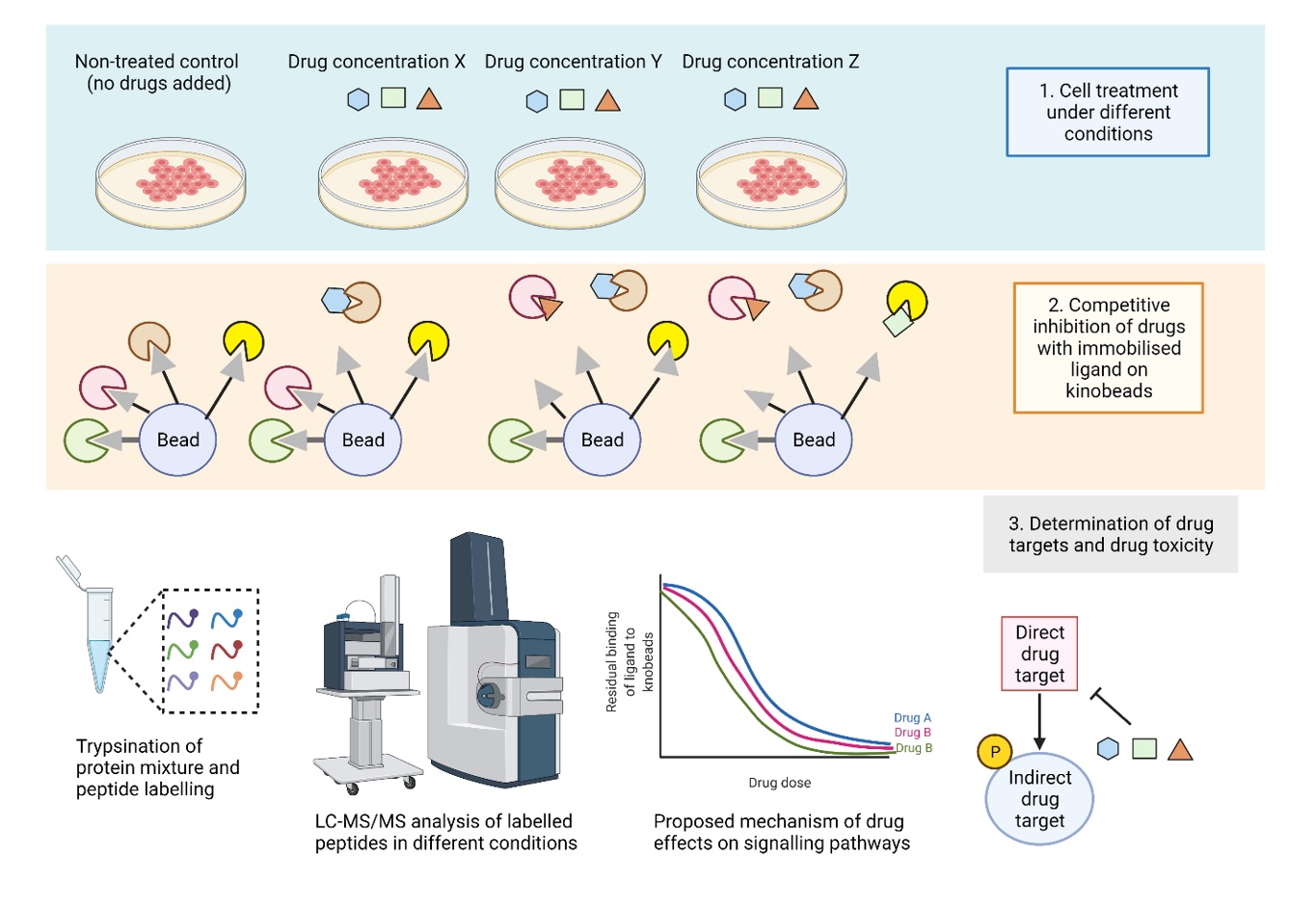
Figure 1. Kinobeads workflow. Cells are pre-incubated with increasing drug concentrations or control together with the kinobeads suspension. The free drug compound and kinobeads compete for the same binding site of the kinase in the cell or tissue lysates, which leads to reduced enrichment of the targets by kinobeads. Proteins are eluted from beads, digested and measured via LC–MS/MS. The binding curves are generated based on the relative intensity values at each drug concentration and non-linear regression analysis (Created with
BioRender.com).
References:
[1] Bhullar KS, Lagarón NO, McGowan EM, Parmar I, Jha A, Hubbard BP, Rupasinghe HPV. Kinase-targeted cancer therapies: progress, challenges and future directions. Mol Cancer. 2018;17(1):48.
[2] Lee PY, Yeoh Y, Low TY. A recent update on small-molecule kinase inhibitors for targeted cancer therapy and their therapeutic insights from mass spectrometry-based proteomic analysis. FEBS J. 2022. doi: 10.1111/febs.16442. Epub ahead of print.
[3] Klaeger S, Heinzlmeir S, Wilhelm M, Polzer H, Vick B, Koenig PA, et al. The target landscape of clinical kinase drugs. Science. 2017;358(6367):eaan4368.
[4] Davis MI, Hunt JP, Herrgard S, Ciceri P, Wodicka LM, Pallares G, Hocker M, Treiber DK, Zarrinkar PP. Comprehensive analysis of kinase inhibitor selectivity. Nat Biotechnol. 2011;29(11):1046-51.
[5] Golkowski M, Brigham JL, Perera GK, Romano GE, Maly DJ, Ong SE. Rapid profiling of protein kinase inhibitors by quantitative proteomics. Medchemcomm. 2014;5(3):363-369.
[6] Anastassiadis T, Deacon SW, Devarajan K, Ma H, Peterson JR. Comprehensive assay of kinase catalytic activity reveals features of kinase inhibitor selectivity. Nat Biotechnol. 2011;29(11):1039-45.
[7] Bantscheff M, Drewes G. Chemoproteomic approaches to drug target identification and drug profiling. Bioorg Med Chem. 2012;20(6):1973-8.
[8] Bantscheff M, Eberhard D, Abraham Y, Bastuck S, Boesche M, Hobson S, et al. Quantitative chemical proteomics reveals mechanisms of action of clinical ABL kinase inhibitors. Nat Biotechnol. 2007;25(9):1035-44.
[9] Knapp S. New opportunities for kinase drug repurposing and target discovery. Br J Cancer. 2018;118(7):936-937.

