microRNA at a glance
Written by: Mira Farzana binti Mohamad Mokhtar & Dr. Tan Shing Cheng
Date published: 23 March 2020
What is microRNA?
The central dogma of molecular biology posits that genetic information flows in a two-step process that comprises transcription (DNA to RNA) and translation (RNA to protein). For decades, scientists had thought that RNA played a minor intermediary role in the entire process. However, starting from the 1960s, research has found that there are many subsets of RNAs which may give an impact to the formation of proteins by mechanisms such as RNA silencing or post-transcriptional regulation of gene expression. One of the most studied subsets of RNA is the microRNA (miRNA), which is a small non-coding RNA usually 21-24 nucleotides long, present in all eukaryotic cells and is highly conserved [1].
The first miRNA, lin-4 (abnormal cell LINeage) was discovered by Victor R. Ambros and co-workers in 1993 when they were studying the developmental process of Caenorhabditis elegans. The single-stranded sequence produces a 22-nucleotide long regulatory RNA rather than coding for a protein. The lin-4 RNA was found to pair with mRNAs involved in the developmental process through complementary base pairing and in this way, functioned as an endogenous regulator of genes [2]. Over the past decades, a number of important milestones have been achieved in the field of microRNA (Figure 1). Victor R. Ambros received the 2008 Albert Lasker Award for Basic Medical Research along with other two scientists, Gary B. Ruvkun and David C. Baulcomber, for their discoveries of microRNA.
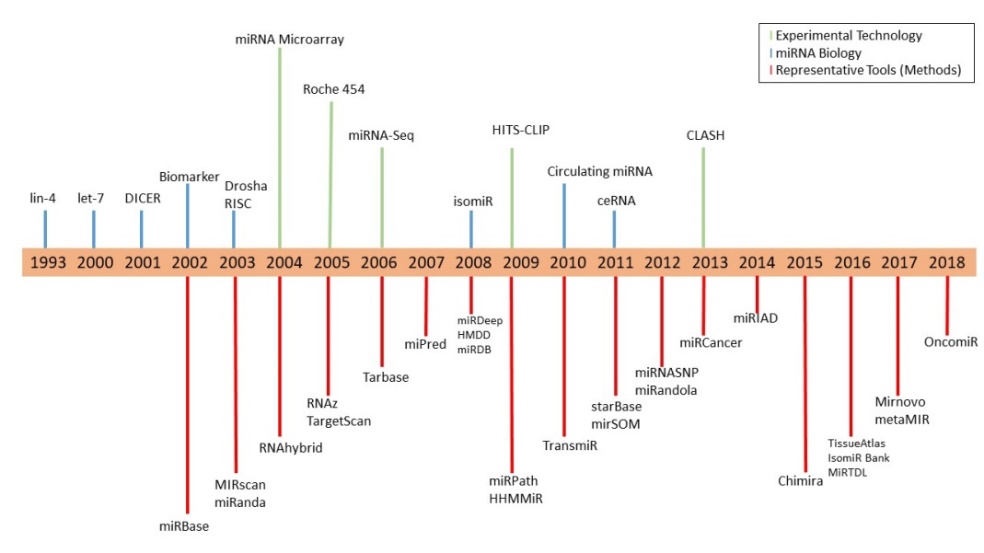
Figure 1. The timeline of miRNA research from 1993 until 2018 (Adapted from [3])
Mechanisms of microRNA action
All microRNAs are produced from their respective genes through a series of steps (Figure 2). They are initially transcribed into hairpin-shaped pri-microRNAs, which are then processed by Drosha enzyme into pre-microRNAs. Pre-microRNAs are subsequently processed by Dicer, resulting in the formation of duplex microRNA strands. One of these strands is then loaded into the RNA induced silencing complex (RISC) where it interacts with mRNA [4]. When the microRNA binds perfectly with mRNA, it will lead to the cleavage of mRNA, but when the microRNA binds only partially with mRNA, it inhibits the mRNA from being translated.
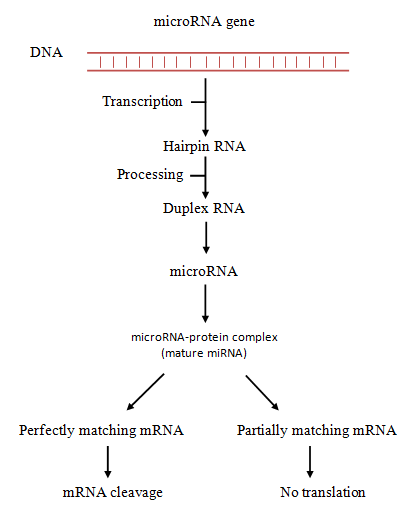
Figure 2. Biogenesis and mechanisms of microRNAs
Relevance of microRNAs in disease
microRNAs have been known to be involved in many biological processes, such as inflammation, as well as apoptosis, metastasis and proliferation [5]. This implicates a role for them in various diseases, notably cancer. In the UKM Medical Molecular Biology Institute (UMBI), a number of research have been carried out to investigate the relevance of microRNA in cancers [6-8].
MicroRNA target prediction
Since microRNAs can regulate a large number of genes, prediction of their target genes can help to identify the pathways in which the microRNA is relevant [9]. Prediction of microRNA targets can be done via computational (bioinformatics/in silico) approach. Some of the target prediction tools available online are DIANA-MicroT, MirPath, TargetScan, MirBase, miRDB and DIANA-tarbase. The latest release of miRBase v22 contains 38,589 entries that represent microRNAs from 271 organism. The microRNA class comprises approximately 2,654 mature miRNAs in human and 469 in D.melanogaster [10]. Nevertheless, there is still a lot more to understand and discover about the precise mechanism of microRNA in specific functions. With the abundance data on microRNA over the past 10 years, it is hoped that the precise impact of microRNA can be fully characterized in near future.
References
1. Romano, G., Veneziano, D., Acunzo, M., & Croce, C. M. (2017). Small non-coding RNA and cancer. Carcinogenesis, 38(5), 485–491. https://doi.org/10.1093/carcin/bgx026
2. Lai, E. C. (2015). Two decades of miRNA biology: Lessons and challenges. Rna, 21(4), 675–677. https://doi.org/10.1261/rna.051193.115
3. Chen, L., Heikkinen, L., Wang, C., Yang, Y., Sun, H., & Wong, G. (2019). Trends in the development of miRNA bioinformatics tools. Briefings in Bioinformatics, 20(5), 1836–1852. https://doi.org/10.1093/bib/bby054
4. Hammond, S.M. (2015). An overview of microRNAs. Adv Drug Deliv Rev. 2015 June 29; 87: 3–14. doi:10.1016/j.addr.2015.05.001
5. Garofalo, Michela & Croce, Carlo. (2011). MicroRNAs: Master Regulators as Potential Therapeutics in Cancer. Annual review of pharmacology and toxicology. 51. 25-43. 10.1146/annurev-pharmtox-010510-100517.
6. Tan, S. C., Lim, P. Y., Fang, J., Mokhtar, M. F. M., Hanif, E. A. M., & Jamal, R. (2020). Association between MIR499A rs3746444 polymorphism and breast cancer susceptibility: a meta-analysis. Scientific Reports (accepted for publication).
7. Sulaiman, S. A., Abu, N., Ab-Mutalib, N. S., Low, T. Y., & Jamal, R (2019). Signatures of gene expression, DNA methylation and microRNAs of hepatocellular carcinoma with vascular invasion. Future Oncology, 15(22), 2603-2617.
8. Hanif, E. A. M. (2019). Dysregulation of non-histone molecule miR205 and LRG1 post-transcriptional de-regulation by SETD1A in triple negative breast cancer. Mol Biol Rep, 46(6), 6617-6624.
9. Tripathi, A., Goswami, K., & Sanan-Mishra, N. (2015). Role of bioinformatics in establishing microRNAs as modulators of abiotic stress responses: The new revolution. Frontiers in Physiology, 6(OCT), 286. https://doi.org/10.3389/fphys.2015.00286
10. Kozomara, A., Birgaoanu, M., & Griffiths-Jones, S. (2019). MiRBase: From microRNA sequences to function. Nucleic Acids Research, 47(D1), D155–D162. https://doi.org/10.1093/nar/gky1141

