Pendekatan Proteomik berasaskan Spektrometri Jisim untuk Analisis Perencat Kinase
-
Ditulis oleh: Profesor Madya Dr. Low Teck Yew, Dr. Lee Pey Yee
Tarikh : 18 Mei 2022
Institut Biologi Molekul Perubatan UKM (UMBI), Universiti Kebangsaan Malaysia, 56000 Kuala Lumpur, Malaysia
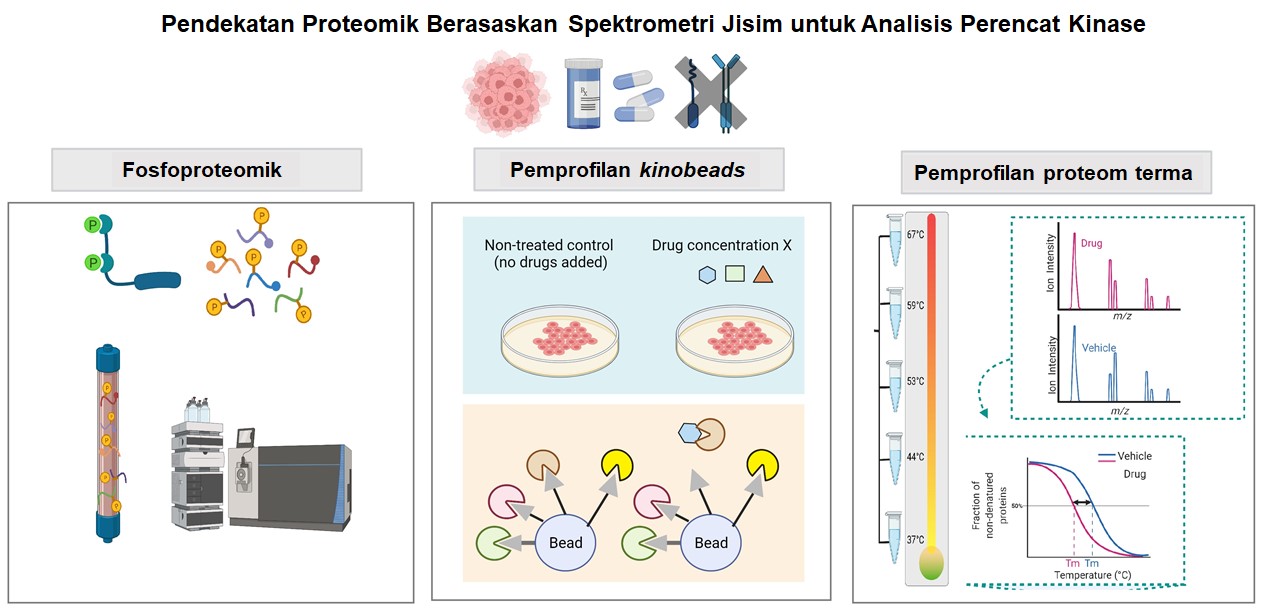
Kinase adalah protein isyarat pengawalseliaan utama yang mengawal pelbagai proses biologi penting dan fungsi selular. Disregulasi kinase protein terlibat dalam permulaan dan perkembangan kebanyakan kanser. Memandangkan peranan penting kinase, terdapat peningkatan usaha dalam memanfaatkan kinase sebagai ubat molekul sasaran untuk rawatan kanser. Bermula dengan kejayaan imatinib (Gleevec) – perencat kinase tirosina pertama yang diluluskan oleh Pentadbiran Makanan dan Ubat-ubatan (FDA) Amerika Syarika pada tahun 2001 yang menyasarkan Bcr-Abl untuk merawat leukemia myelogenous kronik (CML), terapi dengan perencat kinase telah berkembang pesat. Dalam beberapa dekad kebelakangan ini, banyak perencat kinase molekul kecil telah dibangunkan dan merevolusikan landskap rawatan kanser. Perencat kinase molekul kecil secara amnya boleh dikategorikan kepada tujuh jenis berdasarkan mekanisme tindakan dan mod pengikatan sasaran. Sehingga April 2022, FDA telah meluluskan 71 perencat kinase molekul kecil untuk kegunaan klinikal (http://www.brimr.org/PKI/PKIs.htm), kebanyakannya adalah untuk rawatan kanser. Walaupun berpotensi besar, terdapat pelbagai cabaran dalam membangunkan perencat kinase yang sangat selektif dan berkesan, seperti isu ketoksikan dan rintangan yang kerap timbul. Dalam konteks ini, aplikasi proteomik berasaskan spektrometri jisim (MS) yang menyediakan profil kandungan protein yang luas boleh membantu proses penemuan dan pembangunan ubat perencat kinase. Pendekatan proteomik berasaskan MS seperti fosfoproteomik, kinobeads dan pemprofilan proteom terma berguna untuk pelbagai aspek penyelidikan perencat kinase. Fosfoproteomik bertujuan untuk mengenal pasti dan mengukur fosfopeptida, serta tapak fosforilasi secara global dengan MS. Fosfoproteomik telah digunakan dalam pembangunan terapeutik untuk menentukan kinase utama dan litar isyarat yang berkaitan dengan tindak balas ubat. Teknologi kinobeads proteomik kimia menggunakan matriks afiniti terhadap kinase untuk menangkap rakan interaksi, diikuti dengan pengenalpastian oleh MS. Teknik ini telah menjadi strategi yang berguna untuk menentukan landskap sasaran perencat kinase dan rangkaian berfungsi. Sebaliknya, pemprofilan protein terma (TTP) menganalisis interaksi ubat-protein dengan menilai perubahan dalam kestabilan protein terhadap denaturasi haba. TPP membolehkan pengenalpastian sasaran ubat dalam lisat sel atau sel hidup dan membenarkan pengukuran perubahan dalam proteom selular akibat interaksi ubat-protein. Oleh itu, TPP berupaya untuk mencirikan tindak balas biologi serta kesan luar sasaran.
Dalam artikel yang baru diterbitkan, kami menerangkan tentang peranan kinase dalam karsinogenesis dan perkembangan terkini dalam perencat kinase molekul kecil yang telah diluluskan untuk kegunaan klinikal. Seterusnya, kami membincangkan pengaplikasian strategi proteomik berasaskan MS dalam kajian perencat kinase. Selain itu, kami juga membincangkan cabaran dan masa depan berkaitan dengan teknik proteomik berasaskan MS untuk penyelidikan perencat kinase.
Rujukan:
Lee PY, Yeoh Y, Low TY. A recent update on small-molecule kinase inhibitors for targeted cancer therapy and their therapeutic insights from mass spectrometry-based proteomic analysis. FEBS J. 2022. doi: 10.1111/febs.16442. Epub ahead of print.
-
Written by: Profesor Madya Dr. Low Teck Yew, Dr. Lee Pey Yee
Date : 18 Mei 2022
UKM Medical Molecular Biology Institute (UMBI), Universiti Kebangsaan Malaysia, 56000 Kuala Lumpur, Malaysia
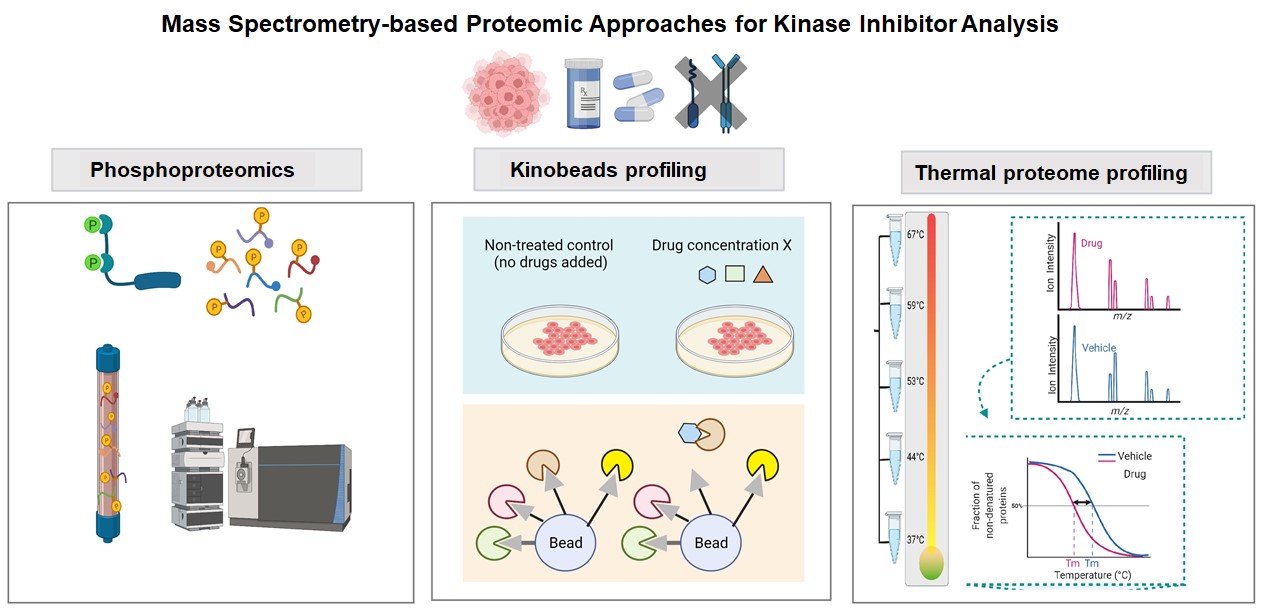
Kinases are key regulatory signalling proteins governing numerous essential biological processes and cellular functions. Dysregulation of many protein kinases is associated with cancer initiation and progression. Given their crucial roles, there has been increasing interest in harnessing kinases as prospective drug targets for cancer. Starting with the success of imatinib (Gleevec) – the first tyrosine kinase inhibitor (TKI) approved by the U.S. Food and Drug Administration (FDA) in 2001 that targets Bcr-Abl for treating chronic myelogenous leukemia (CML), kinase-targeting drugs are burgeoning. In recent decades, numerous small-molecule kinase inhibitors (KIs) have been developed and revolutionized the cancer treatment landscape. Small molecule KIs can be generally categorized into seven types based on their mechanisms of action and modes of target binding. As of April 2022, the FDA has approved 71 small molecule kinase inhibitors for clinical usage (http://www.brimr.org/PKI/PKIs.htm), many of which are for cancer treatment. Despite their great potential, challenges remain in developing highly selective and effective kinase inhibitors, with toxicity and resistance issues frequently arising. In this context, the application of MS-based proteomics which provides unbiased profiling of protein content could offer important insights that facilitate the kinase drug discovery and development process. MS-based proteomics approaches such as phosphoproteomics, kinobeads and thermal proteome profiling are useful for various aspects of kinase drug research. Phosphoproteomics aims to identify and quantify phosphopeptides, as well as the phosphorylation sites in a global, high-throughput and specific manner with MS. Phosphoproteomics has been applied in therapeutic development for unravelling key kinases and signalling circuitry associated with drug response. The chemical proteomics kinobeads technology utilizes an affinity matrix for kinases to capture binding partners, followed by identifying the bound proteins by MS. This technique has become a useful strategy for defining the target landscape of KIs and the functional networks. Thermal proteome profiling (TTP), on the other hand, analyze drug-protein interactions by assessing changes in the protein stability upon thermal denaturation. TPP is based on the principle that the thermal stability of proteins is altered upon interaction with a ligand, causing the proteins to become more resistant to heat-induced aggregation. TPP enables the identification of direct and indirect drug targets in cell lysates or living cells and allows the measurement of changes in the cellular proteome upon drug-protein interactions. As such, TPP provides the capability to characterize the biological response of the on-target engagement as well as the downstream effects of the off-target binding.
In a recent review, we provide an overview of the role of kinases in carcinogenesis and describe the current progress with small-molecule kinase inhibitors that have been approved for clinical use. We then discuss the application of mass spectrometry (MS)-based proteomics strategies in the study of kinase inhibitors. Finally, we discuss the challenges and outlook concerning MS-based proteomics techniques for kinase drug research.
Reference:
Lee PY, Yeoh Y, Low TY. A recent update on small-molecule kinase inhibitors for targeted cancer therapy and their therapeutic insights from mass spectrometry-based proteomic analysis. FEBS J. 2022. doi: 10.1111/febs.16442. Epub ahead of print.

