Workshop Outline
This introductory workshop will focus on:
-
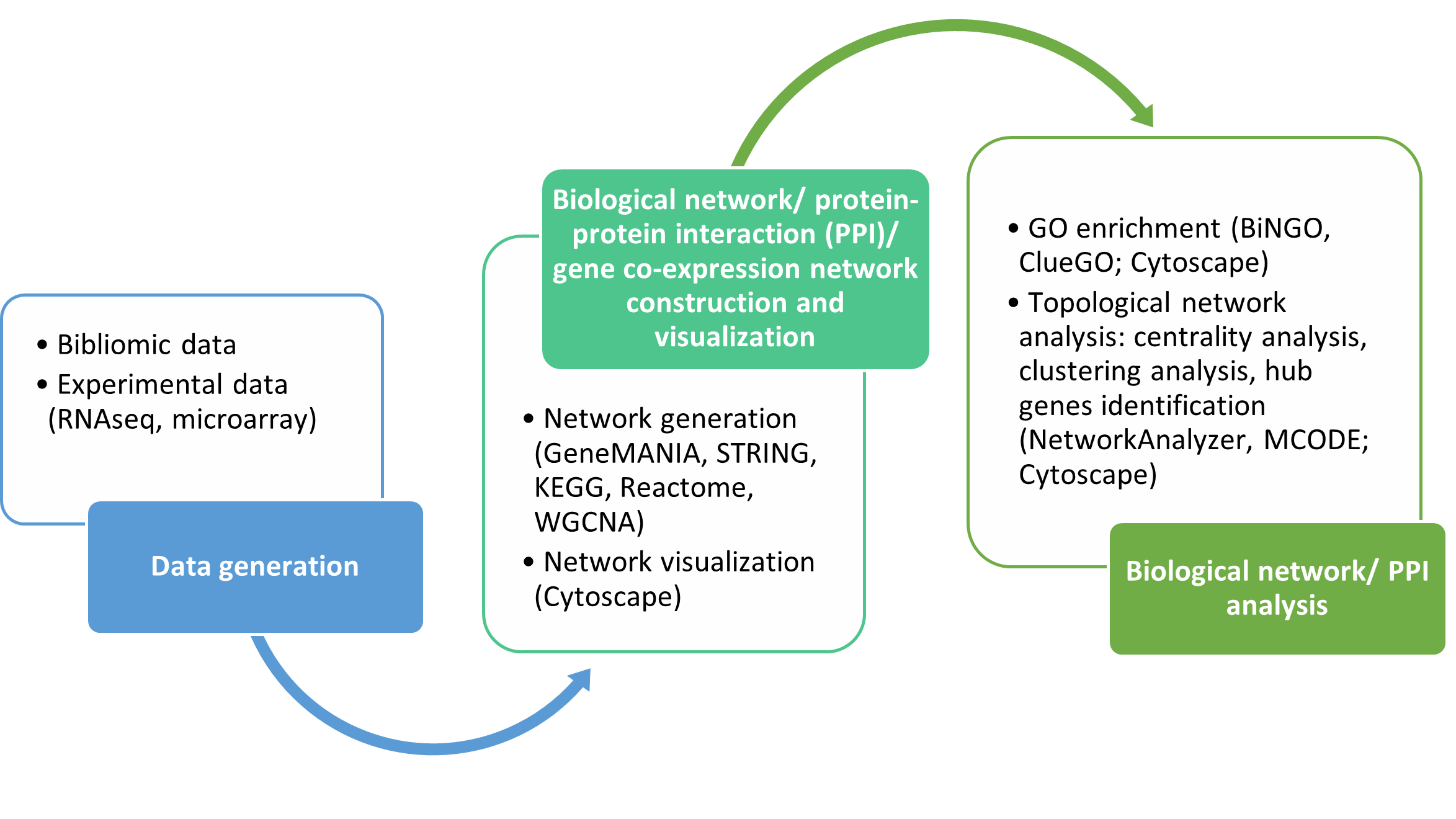
Construction of protein-protein interaction (PPI) using relevant databases i.e., STRING and GeneMANIA
-
Mapping of identified genes onto KEGG pathways
-
Construction of gene co-expression network using R package for gene co-expression network construction (WGCNA)
-
Functional annotation of genes/ proteins
-
Gene ontology (GO) enrichment of genes/ proteins
-
Pathway analysis and visualization
Upon the completion of this workshop, participants will be able to:
-
Construct pathways and PPI network from the list of genes/ proteins
-
Construct gene co-expression network using WGCNA
-
Perform GO and pathway analysis on the constructed pathway and PPI network
-
Understand a complete workflow of data acquisition, pathway construction, pathway analysis, and data interpretation.
Who should attend
Anyone interested in pathway analysis and its applications (researchers, academicians, and students), especially biologists from beginner to advanced levels.
List of Speakers and Trainers

-
Dr. Sarahani Harun is currently a research fellow at the Institute of Systems Biology (INBIOSIS). She graduated from the Universiti Kebangsaan Malaysia where she earned a Bachelor in Bioinformatics, Master in Molecular Biology and PhD in Systems Biology. Her research interest involves computational biology that includes pathway analysis in secondary metabolites, i.e., glucosinolates and cyanogenic glycosides to unravel potential genes in the respective biosynthetic pathways. This computational pipeline can be used for new gene discovery to establish genetic resources for crop improvement.
-
She is also interested in analyzing the transcriptome data generated from the oil pest, bagworm (Metisa plana) which is a collaborative project with Felda Global Ventures R&D. The identification of the key regulatory genes from the study would facilitate in controlling the pest infestation in oil palm. She is currently the coordinator for the Centre of Omics Data Analysis (CODA) (https://www.ukm.my/inbiosis/en/coda/). CODA is a one-stop solution research centre for omics data analysis. This centre provides consultation and assistance on high-throughput data analysis that is associated with systems biology.
-
Dr. Sarahani can be contacted via email; sarahani@ukm.edu.my.
-
Dr. Nisha Govender is a research fellow at the Institute of Systems Biology, UKM. She earned her professional degrees at UKM and UPM in the field of plant sciences. Nisha’s research interests are mainly in the field of plant disease, breeding and nutraceuticals of oil seed crops (oil palm, Jatropha), food crops (rice, jackfruit, breadfruit) and herbal plants (whisker’s cat, pea eggplant, Mexican mint, water dropwort).
-
Her current research activities are indicated as following: i) green synthesis of nanosilica fertilizer for soil fertility enhancement, ii) identification of natural bioactive compounds via molecular docking and molecular dynamic simulation studies on Malaysian herbal plants, and iii) Identification of RNAi components in Ganoderma boninense, the causal agent of basal stem rot of oil palm. If you have a parallel interest, wish to learn more, or/and interested to participate in her research, feel free to write to nishag@ukm.edu.my.


-
Prof. Dr. Zeti Azura Mohamed Hussein is a Professor in Bioinformatics at the Department of Applied Physics, Faculty Science and Technology. She investigates the relationships between disease genes and disease networks using gene network analysis, pathway analysis and network analysis to understand disease pathobiology in human and plants alike which can’t be fully annotated using a single type of data thus the inclusion of big data analysis and omics data integration. Prof. Zeti’s research demonstrates how integrative analysis on omics data can be used to unravel new important knowledge and provide answers on the biological problems of interest. If you are interested to collaborate with her, or be part of her research group, please drop her an email to zeti.hussein@ukm.edu.my or visit her website at https://zetibioinfolab.wordpress.com
Programme
19th September 2023 (Tuesday)
| Time | Activity | Trainer |
| 8.30 am | Registration | - |
| 9.00 am | Introduction | Dr. Sarahani Harun |
| 9.10 am | Lecture 1: Introduction to pathway and biological network | Prof. Dr. Zeti Azura Mohamed Hussein |
| 10.30 am | Tea break | - |
| 11.00 am | Practical 1: Introduction to network analysis, databases & tools (Cytoscape);
Retrieving pathway/ networks from databases |
Dr. Sarahani Harun |
| 12.30 pm | Lunch break | - |
| 2.00 pm | Practical 2 : Pathway / network visualization;
Editing pathway (layout, style) |
Dr. Sarahani Harun |
| 4.00 pm | Afternoon break | - |
| 4.15 pm | Summary, discussions and wrap-up | Dr. Sarahani Harun |
| 5.00 pm | END OF DAY 1 |
|
20th September 2023 (Wednesday)
| Time | Activity | Trainer |
| 9.00 am | Introduction | Dr. Sarahani Harun |
| 9.10 am | Lecture 2: Gene co-expression network analysis | Dr. Sarahani Harun |
| 10.30 am | Tea break | - |
| 11.00 am | Practical 3: Introduction to R basics - (installation & download) | Dr. Nisha T. Govender |
| 12.30 pm | Lunch break | - |
| 2.00 pm | Practical 4: Hands-on session using R package for gene co-expression network construction (WGCNA) | Dr. Nisha T. Govender |
| 4.00 pm | Afternoon break | - |
| 4.15 pm | Summary, discussions and wrap-up | - |
| 5.00 pm | END OF DAY 2 |
|
21st September 2023 (Thursday)
| Time | Activity | Trainer |
| 9.00 am | Introduction to Day 3 & Recap | - |
| 9.10 am | Lecture 3: Introduction to network analysis and data integration | Dr. Sarahani Harun |
| 10.30 am | Morning break | - |
| 10.45 am | Practical 5: Cytoscape – extending pathways/ networks (merging pathways/ networks) | Dr. Sarahani Harun |
| 1.00 pm | Lunch break | - |
| 2.00 pm | Practical 6 : Cytoscape – GO enrichment/ KEGG pathway analysis | Dr. Sarahani Harun |
| 4.00 pm | Afternoon break | - |
| 4.15 pm |
|
- |
| 5.00 pm | END OF WORKSHOP |
|

